Using Kale + Kubeflow Katib to Save Best Fitted Model
Saving each fitted model and parameters for hyperparameter tuning with Kubeflow Katib.
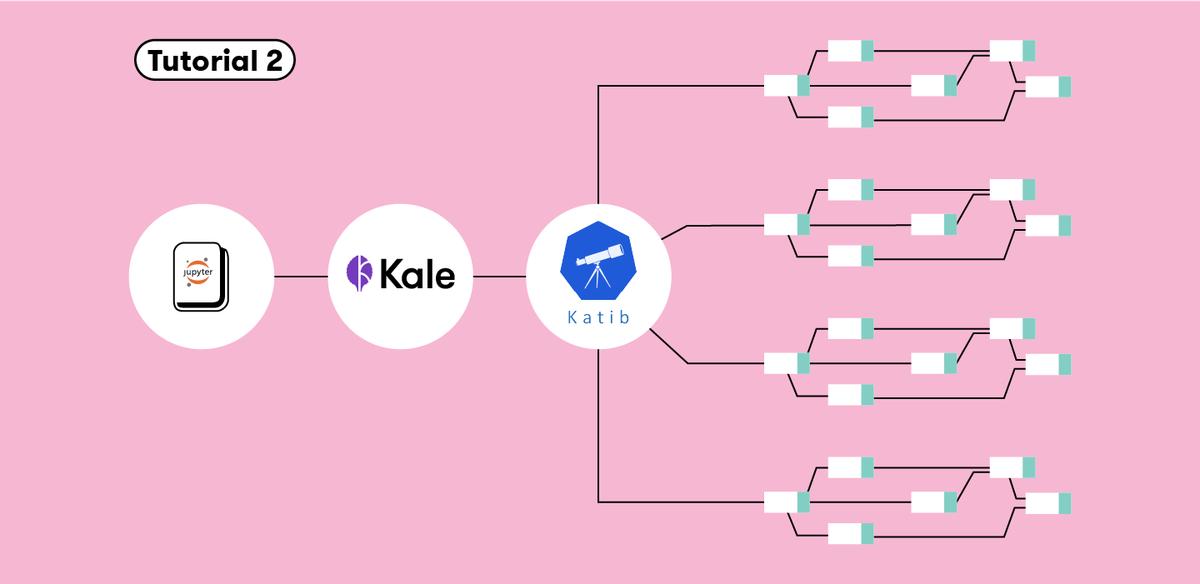 Build An End-to-End ML Workflow: From Notebook to HP Tuning to Kubeflow Pipelines with Kale
Build An End-to-End ML Workflow: From Notebook to HP Tuning to Kubeflow Pipelines with Kale
Overview
Kale can be used to unify the workflow across components of ML projects, and presents a seamless process to create ML pipelines for HP tuning, starting from your Jupyter Notebook. One can use Kale to convert a Jupyter Notebook to a Kubeflow Pipeline without any modification to the original Python code. Within the Kale panel we can enable Hyperparameter tuning with Kubeflow Katib.
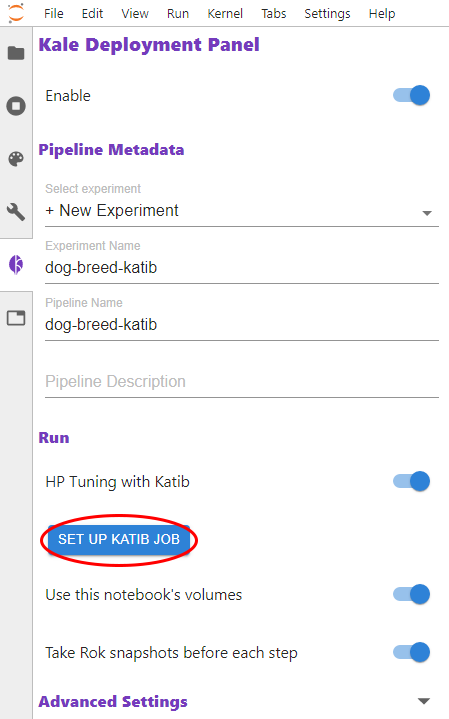
Eventually, Kubeflow Katib will return information about the the best fitted model to the user.
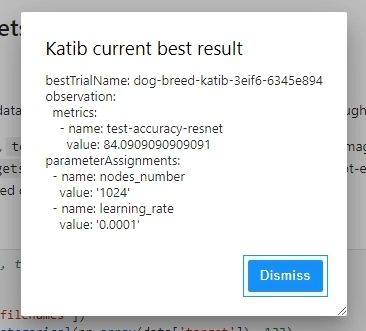
While metadata gets logged during pipeline execution, there were some limitations that I encountered when using Kubeflow Katib.
- When using Katib, there is no apparent way to store fitted models/parameters in a location/registry
- E.g. It would be useful for us to know the name of the best trained model, parameters, and its model location in a storage bucket.
One workaround is to use ‘kubectl’ commands to try and save fitted models and parameters in some location (e.g. storage bucket). Disclaimer: This workaround is cumbersome, and requires running Katib trials sequentially, which is less than optimal. But hey – it works for now (until I figure out a better solution)! Some code below to do this:
storage_client = storage.Client()
model_filename = 'model.pkl'
with open(model_filename, 'wb') as f:
pickle.dump(dct, f)
# save model params
params_filename = 'params.pkl'
params={'params' : dct.get_params(),'feature_names' : train.columns.to_list()}
with open(params_filename, 'wb') as f:
pickle.dump(params, f)
resul = os.popen('kubectl get experiment -n kubeflow-user').readlines()
res = [resul[x].split() for x in range(len(resul))]
res=pd.DataFrame(res[1:], columns=res[0])
experiment_name = res[res.STATUS=='Running']['NAME'].values[0]
bucket = storage_client.get_bucket('bm-kubeflow-lab-kubeflowpipelines-default')
resul = os.popen('kubectl get trial -n kubeflow-user').readlines()
res = [resul[x].split() for x in range(len(resul))]
res=pd.DataFrame(res[1:], columns=res[0])
trial_name = res[res.TYPE=='Running']['NAME'].values[0]
model_load = bucket.blob('test_upload/{}/{}/{}'.format(experiment_name, trial_name,model_filename))
model_load.upload_from_filename(model_filename)
params_load = bucket.blob('test_upload/{}/{}/{}'.format(experiment_name, trial_name,params_filename))
params_load.upload_from_filename(params_filename)
#store experiment name in bigquery table to track experiment names, and dates in which jobs were run.
exp_table = pd.DataFrame(data=[experiment_name], columns=['experiment_name'])
bq_client = bq.Client()
dataset_ref = bq_client.dataset('project_dataset')
table_ref = dataset_ref.table('katib_experiments')
job_config = bq.LoadJobConfig()
job_config.write_disposition = bq.WriteDisposition.WRITE_TRUNCATE
job = bq_client.load_table_from_dataframe(dataframe=exp_table, destination=table_ref, job_config=job_config)
job.result()
After hyperparameter tuning has run sucessfully with Kubeflow Katib, we can access the name of the best trial, go into its associated directory where the fitted model is stored and move this to a separate directory for later access (e.g. for serving scripts)
# check best model and update a seperate "deploy" folder with this model. Might need to put this source code as a final step executed after full pipeline is run.
resul=os.popen('kubectl -n kubeflow-user get experiment {} -o yaml'.format(experiment_name)).readlines()
res =[resul[x].split() for x in range(len(resul))]
res=pd.DataFrame(res)
best_trial = res[res[0]=='bestTrialName:'][1].values[0]
if best_trial != '""':
files = bucket.list_blobs(prefix='test_upload/{}/{}/'.format(experiment_name ,best_trial))
fileList = [file for file in files]
for file in fileList:
file_name = file.name.split('/')[-1]
bucket.rename_blob(file, 'test_upload/best_models/deploy/{}'.format(file_name))